Top Qs
Timeline
Chat
Perspective
Pyrimidine dimer
Type of damage to DNA From Wikipedia, the free encyclopedia
Remove ads
Main article: Pyrimidine
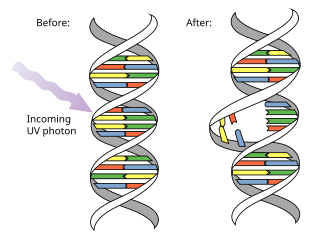
A pyrimidine dimer is a type of molecular lesion that arises when adjacent thymine or cytosine bases are bonded together in an atypical way, often as a result of a photochemical reaction.[1][2] Ultraviolet light (UV),[3] particularly UVC, often causes this direct DNA damage, causing the formation of covalent bonds near the nucleotides' carbon–carbon double bonds.[4] The resulting photo-coupled dimers are fluorescent,[5] and are commonly classified as cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These pre-mutagenic lesions modify the DNA helix structure by distorting it.
Up to 100 dimerization reactions per second can occur in a skin cell exposed to sunlight,[6] resulting in DNA damage. However, these lesions are typically rectified by DNA repair mechanisms before they can cause lasting damage. These mechanisms include photolyase reactivation and nucleotide excision repair, the latter of which is prevalent in humans. A failure to quickly repair these lesions may lead to erroneous (non-canonical) nucleotide incorporation by polymerase machinery.[7] Extreme DNA damage can precipitate mutations within an organism's genome, potentially culminating in cancer cell formation.[8] These lesions may also interfere with polymerase function, induce transcription or replication errors, or halt replication.[9]
Pyrimidine dimers contribute to sunburn and melanin production, and are a primary factor in melanoma development in humans.[10]
Remove ads
Types of pyrimidine dimers

The different types of pyrimidine dimers each have distinct structures and implications for DNA integrity.
A cyclobutane dimer (CPD) features a four-membered ring formed by the fusion of two double-bonded carbons from adjacent pyrimidines. CPDs disrupt the formation of the base pair during DNA replication, which can potentially lead to mutations.[11][12][13]
The 6–4 photoproduct (6–4 pyrimidine–pyrimidone, or 6–4 pyrimidine–pyrimidinone) is an alternate dimer configuration which covalently links the carbon at the 6 (C6) position of one pyrimidine ring and the carbon at the 4 (C4) position of the adjoining base's ring.[14] This type of conversion occurs at one third of the frequency of CPDs but has a higher mutagenic risk.[15]
A Dewar pyrimidinone results from the reversible isomerization of a 6–4 photoproduct under further light exposure.[16]
Remove ads
Mutagenesis
Mutagenesis, the process of mutation formation, is significantly influenced by translesion polymerases which often introduce mutations at sites of pyrimidine dimers.[17] This occurs in prokaryotes through the SOS response to mutagenesis and in eukaryotes through other methods. As thymine–thymine CPDs are the most common lesions induced by UV, translesion polymerases show a tendency to incorporate adenines opposite these dimers, resulting in accurate replication. Cytosines that are part of CPDs, however, are susceptible to deamination, leading to cytosine to thymine transitions and contributing to the mutation process.[18]
Remove ads
DNA repair
Summarize
Perspective

Pyrimidine dimers introduce local conformational changes in the DNA structure, which allows recognition of the lesion by repair enzymes.[19] In most organisms (excluding placental mammals such as humans) they can be repaired by photoreactivation.[20] Photoreactivation is a repair process in which photolyase enzymes reverse CPDs using photochemical reactions. In addition, some photolyases can also repair 6-4 photoproducts of UV induced DNA damage. Photolyase enzymes utilize flavin adenine dinucleotide (FAD) as a cofactor in the repair process.[21]
The UV dose that reduces a population of wild-type yeast cells to 37% (assuming a Poisson distribution of hits) is the same as the UV dose that causes an average of one lethal hit to each of the cells of the population.[22] The number of pyrimidine dimers induced per haploid genome at this dose was measured as 27,000.[22] A mutant yeast strain defective in the three known pyrimidine dimer repair pathways was also tested for UV sensitivity. In this case, only one to two unrepaired pyrimidine dimers per haploid genome are lethal to the cell.[22] These findings thus indicate that the repair of thymine dimers in wild-type yeast is highly efficient.[citation needed]
Nucleotide excision repair (NER), sometimes termed "dark reactivation", is a more general mechanism for repair of lesions and is the most common form of DNA repair for pyrimidine dimers in humans. This process works by using cellular machinery to locate the dimerized nucleotides and excise the lesion. Once the CPD is removed, there is a gap in the DNA strand that must be filled. DNA machinery uses the undamaged complementary DNA strand as a template to synthesize the matching nucleotides and consequently fill in the gap on the damaged strand.[9]
Xeroderma pigmentosum (XP) is a rare genetic disease in humans that is caused by UV damage to genes that code for NER proteins, resulting in the inability for the cell to combat pyrimidine dimers that form. Individuals with XP are also at a much higher risk of cancer, with a >5,000-fold increased risk of developing skin cancers compared to the general population.[8] Some common features and symptoms of XP include skin discoloration and the formation of multiple tumors proceeding UV exposure.[23]
A few organisms have other ways to perform repairs:
- Spore photoproduct lyase is found in spore-forming bacteria. It reverts thymine dimers to their original state.[24]
- Deoxyribodipyrimidine endonucleosidase is found in bacteriophage T4. It is a base excision repair enzyme specific for pyrimidine dimers, and is able to cut open the AP site.
Another type of repair mechanism that is conserved in humans and other non-mammals is translesion synthesis. Typically, the lesion associated with the pyrimidine dimer blocks cellular machinery from synthesizing past the damaged site. However, in translesion synthesis, translesion polymerases can replicate past the CPD, allowing both replication and transcription machinery to continue past the lesion. One specific translesion DNA polymerase, DNA polymerase η, is deficient in individuals with Xeroderma pigmentosum.[25]
Remove ads
See also
References
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads
