Top Qs
Timeline
Chat
Perspective
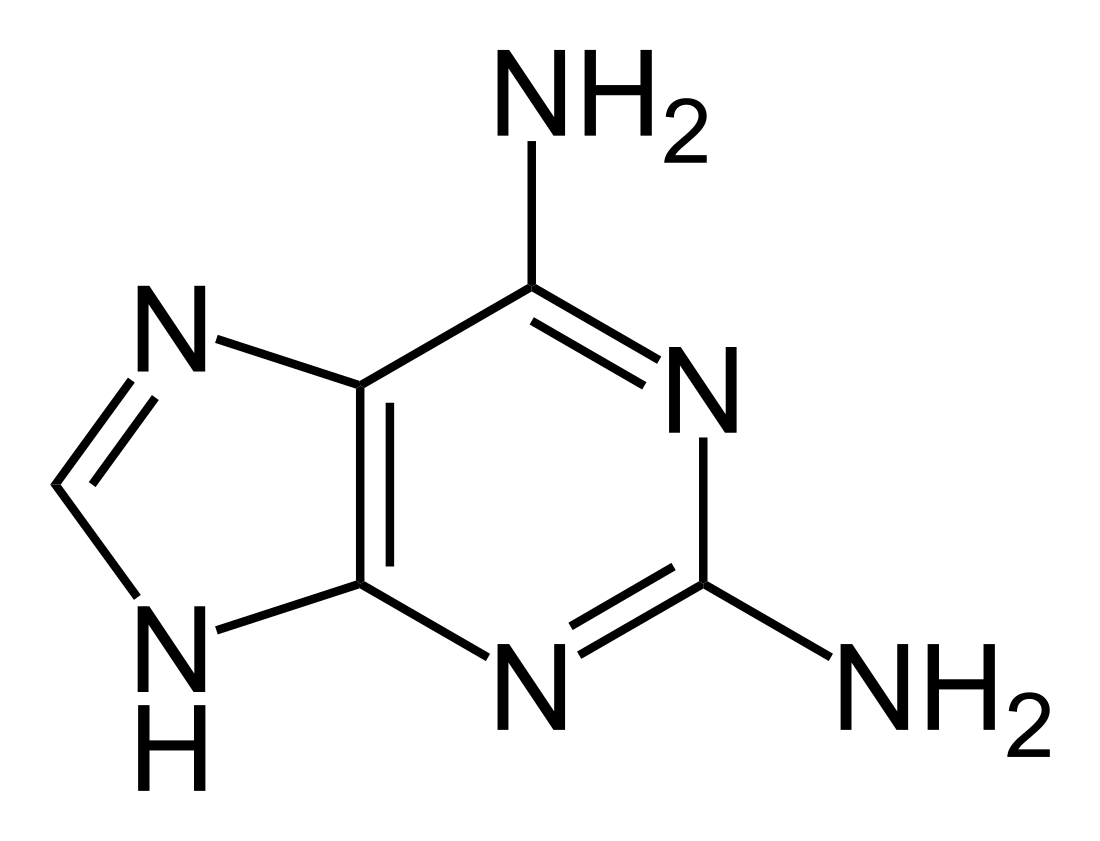
2,6-Diaminopurine
Chemical compound From Wikipedia, the free encyclopedia
Remove ads
2,6-diaminopurine (2,6-DAP, also known as 2-aminoadenine, standard IUPAC symbol n2A[1]: N4.1, N4.4 ) is a compound once used in the treatment of leukemia.[2] It is found instead of adenine (A) in the genetic material of some bacteriophage viruses,[3]
In August 2011, a report, based on NASA studies with meteorites found on Earth, was published suggesting 2,6-diaminopurine and related organic molecules, including the DNA and RNA components adenine and guanine, may have been formed extraterrestrially in outer space.[4][5][6]
Remove ads
Abbreviations
In virology it is referred to as the "Z" base.[7] However, "Z" refers instead to 6-amino-5-nitropyridin-2-one in the Artificially Expanded Genetic Information System (AEGIS), so context is required to differentiate.[8] AEGIS refers to this base as "AminoA" instead.[9]
It has also been called the "D" base,[10][11] but the more common interpretation of "D" is dihydrouridine.[12]
In viruses
Summarize
Perspective

In cyanophage S-2L (Siphoviridae), diaminopurine is used instead of adenine (host evasion).[13] Diaminopurine base (Z) pairs perfectly with thymine (T) as it is identical to adenine (A) but has an amine group at position 2 forming 3 intermolecular hydrogen bonds, eliminating the major difference between the two types of basepairs (weak:A-T and strong:C-G). This improved stability affects protein-binding interactions that rely on those differences.
Four papers published April 2021 further describes the use and production of the Z-base. It is now known that:[14]
- The S-2L phage avoids incorporating A bases in the genome by hydrolyzing dATP (DatZ enzyme);[15]
- The Z base is produced by a pathway involving DUF550 (MazZ) and PurZ in S-2L and Vibrio phage PhiVC8;[7]
- The PrimPol/AEP DNA polymerase responsible for handling the Z base occurs in the same gene cluster as the three aforementioned enzymes;[16]
- The Z base is quite widespread in both Siphoviridae and Podoviridae, based on the occurrence of the said gene cluster.[17]
In August 2021, it was shown that DatZ, MazZ and PurZ are sufficient to replace some occurrence of A by Z in the bacterial genome of E. coli; expression of this system is toxic to the cell. The structures of MazZ (subtype 2) and PurZ are also determined, showing a possible link between PurZ and archaeal versions of PurA.[18]
Remove ads
Biosynthesis
2-aminoadenine is produced in two steps. The enzyme MazZ (homologous to MazG, EC 3.6.1.8) first performs:[18]
- dGTP + H2O = dGMP + diphosphate
The enzyme PurZ (homologous to PurA, EC 6.3.4.4) then performs:[7]
- (d)ATP + dGMP + L-aspartate = (d)ADP + phosphate + 2-aminodeoxyadenylosuccinate (dSMP)
The resulting dSMP is processed by host enzymes analogously to adenylosuccinate to produce dZTP.
In cellular life
This article is missing information about results of the altered H-bond strength in DNA and RNA. (October 2021) |
2,6-DAP was used to treat leukemia since as early as 1951.[19] It is known to arrest progression of cell cycle in mouse leukemia cells by 1989.[20] Cancer cells are known to become resistant to DAP by losing their adenine phosphoribosyltransferase (APRT) function,[21] a process shared with E. coli.[22]
DAP derivatives are in vitro antivirals useful against pseudorabies virus, a economically important livestock disease.[23] This base, in its free form, is able to correct UGA nonsense mutations by encouraging translational readthrough, through the inhibition of FTSJ1.[24]
Remove ads
Bioengineering
In bioengineering, anti-miRNA oligonucleotides (specifically, the serinol nucleic acid [SNA] type) incorporating base Z instead of A show enhanced binding to RNA.[25]
DAP is used similarly to other nuclear acid analogues in the investigation of enzyme structures and mechanisms.[26]
References
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads