Top Qs
Timeline
Chat
Perspective
Artificially Expanded Genetic Information System
Synthetic DNA analog experiment From Wikipedia, the free encyclopedia
Remove ads
Artificially Expanded Genetic Information System (AEGIS) is a synthetic DNA analog experiment that uses some unnatural base pairs from the laboratories of the Foundation for Applied Molecular Evolution in Gainesville, Florida, especially the Steven A. Benner lab. AEGIS is a NASA-funded project to try to understand how extraterrestrial life may have developed.[1] In a 2024 article from the same laboratory, the concept has been broadened into anthropogenic evolvable genetic information systems, still with the same acronym.[2]
Hachimoji DNA and Hachimoji RNA (from Japanese 八文字 hachimoji, "eight letters") is a strict subset of this system and comes from the same laboratory.[3] This leads to four allowed base pairs: two unnatural base pairs formed by the synthetic nucleobases in addition to the two normal pairs. Hachimoji bases have been demonstrated in both DNA and RNA analogs, using deoxyribose and ribose respectively as the backbone sugar.[3][4][5][6][7]
Benefits of such a nucleic acid system may include an enhanced ability to store data, as well as insights into what may be possible in the search for extraterrestrial life.[7][8]
Hachimoji DNA and AEGIS comes from the same team lead by ex-Harvard University chemist Steven Benner and belong to the same NASA funding project.[9][10][11][12][13]
Remove ads
Normal DNA
Summarize
Perspective

Natural DNA is a molecule carrying the genetic instructions used in the growth, development, functioning, and reproduction of all known living organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids; alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life. DNA is a polynucleotide as it is composed of simpler monomeric units called nucleotides; when double-stranded, the two chains coil around each other to form a double helix.[14][15]
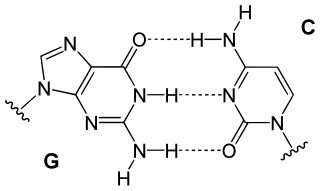
In natural DNA, each nucleotide is composed of one of four nucleobases (cytosine [C], guanine [G], adenine [A] or thymine [T]), a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are bound to each other with hydrogen bonds, according to base pairing rules (A with T and C with G), to make double-stranded DNA.
Remove ads
Bases
Summarize
Perspective
Hachimoji DNA is similar to natural DNA but differs in the number, and type, of nucleobases.[3][7] Unnatural nucleobases, more hydrophobic than natural bases,[16][17] are used in successful hachimoji DNA. Such a DNA always formed the standard double helix, no matter what sequence of bases were used. An enzyme (T7 polymerase) was adapted by the researchers to be used in vitro to transcribe hachimoji DNA into hachimoji RNA, which, in turn, produced chemical activity in the form of a glowing green fluorophore.[6][7]
The full AEGIS system uses 12 to 14 different nucleobases in its genetic code, adding four types of base pairs on top of the two natural Watson-Crick base pairs.[1][18][19][20][12]
Names such as "pyADA", "puDAD" belong to an AGEIS-specific system of denoting nucleobases. PyADA means that the base is a pyrimidine, and from top (5') to down (3') the hydrogen-bonding behavior is acceptor, donor, acceptor. PuDAD means the base is a purine with donor-acceptor-donor pattern. Under this system, all pairs form three hydrogen bonds.[21]
The three-bond system contains considerable flexibility for further modification of nucleosides. Functional groups can be added, removed, or replaced on the non-bonding side of the nucleobase without affecting bonding, much like how uridine (thymine without a methyl group) bonds like thymine in the natural genetic system. The original (1998) formulation only anticipated the possibility of replacing groups on pyAAD (dS), pyADA (T), pyADD (Z), pyDAA (V), and pyDAA (C),[21] but in Benner's 2014 paper all twelve types of bases have one site for group replacement.[22] Benner also indicates in a 2012 report for the DITC that all six purine bases have a second site for attaching another functional group.[23]
In addition, the purine and pyrimidine rings can be replaced with any ring with a similar size. This may prove to be advantageous when reduced tautomerization or some other form of additional stability is needed. For example, 6-amino-5-benzyl-3-methylpyrazin-2-one can replace isoguanine.[24]
Remove ads
Non-canonical bonding
Like natural nucleobases, AEGIS bases can form non-canonical bonds. For example, B can pair with T by tautomerization, Z can pair with G at low pH (protonated hydrogen bond), and P can pair with C at low pH (also protonated). A DNA polymerase without access to the unnatural nucleobases would perform these pairings, causing bases to be replaced.[22]
An analogue of the G·U wobble base pair can be formed between aminoA and 5-methylisocytidine (m5iC).[25]
In 2021, it was found that isoguanine (B) can also base-pair with guanine (G) and 5-aza-7-deazaguanine (P) when put in DNA. The purine-purine base pair requires more space than the typical purine-pyrimidine base pair (the natural Watson-Crick A-T C-G pairs and the designed P-Z B-S pairs are all of these type), but the large groove of the DNA double helix provided enough space for this to happen. This "wider" base pair actually enhances the stability of DNA.[26]
Hoogsteen base pairing results in the formating of triplets in nucleic acid tertiary structure.[27]
Biology
Lack of self-sustainability
Scripps Research chemist Floyd Romesberg, noted for creating the first Unnatural Base Pair (UBP), and expanding the genetic alphabet of four letters to six in 2012,[28] stated that the invention of the hachimoji DNA system is an example of the fact that the natural bases (G, C, A and T) "are not unique".[29][30] Creating new life forms may be possible, at least theoretically,[16] with the new DNA system.[30] For now, however, the hachimoji DNA system is not self-sustaining; the system needs a steady supply of unique building blocks and proteins found only in the laboratory. As a result, "Hachimoji DNA can go nowhere if it escapes the laboratory."[6]
Ribozyme
The hachimoji DNA system produced one type of catalytic RNA (ribozyme or aptamer) in vitro.
Remove ads
Applications
NASA funded this research to "expand[s] the scope of the structures that we might encounter as we search for life in the cosmos".[3] According to Lori Glaze of the Planetary Science Division of NASA, "Life detection is an increasingly important goal of NASA's planetary science missions, and this new work [with hachimoji DNA] will help us to develop effective instruments and experiments that will expand the scope of what we look for."[5][31] Research team leader Steven Benner notes, "By carefully analyzing the roles of shape, size and structure in hachimoji DNA, this work expands our understanding of the types of molecules that might store information in extraterrestrial life on alien worlds."[32]
According to researchers,[3] hachimoji DNA could also be used "to develop clean diagnostics for human diseases, in DNA digital data storage, DNA barcoding, self-assembling nanostructures, and to make proteins with unusual amino acids. Parts of this hachimoji DNA are already being commercially produced by Firebird Biomolecular Sciences LLC".[3][6]
Remove ads
History
Summarize
Perspective
The Benner Lab reported in 1987 that it had expanded the number of bases available for making RNA in an article about the RNA world hypothesis. It is unclear whether the whole AEGIS six-pair arrangement had been planned out by then because the text was not accessible to the author of this article.[33]
The Benner Lab reported in 1989 that the T7 RNA polymerase and the Klenow fragment, in their unmodified forms, can insert an isoguanine (iG) residue when guided by a DNA template containing isocytidine (iC). The study was funded by the Swiss government and Sandoz AG, not by NASA.[34]
A 1990 Benner Lab article reports that T7 RNA polymerase and the Klenow fragment are able to insert 2,4-diaminopyrimidine (kappa, κ) opposite xanthosine (X). The T7 RNA polymerase could insert X opposite of κ but the Klenow fragment could not. This research was also funded by Swiss organizations. It also includes the six-base-pair arrangement of AEGIS, unlike the earlier article.[35]
A 1992 cooperation with Jim Bain and Dick Chamberlin (UC Irvine) produced an example of pyAAD in mRNA and puDDA in tRNA. The result can be translated using a standard ribosome to incorporate the non-standard amino acid iodotyrosine. When the tRNA is absent, the ribosome exhibits a frame shift and continues translating. This is the first study with any funding provided by the US government as one of the authors (Christopher Switzer of UC Riverside) was funded by a NSF fellowship.[36]
A 1998 article from the Benner Lab was funded by the Swiss National Science Foundation, the Danish Natural Science Research Council, the National Institutes of Health, and the Office of Naval Research. It provides an overview of Benner's research so far.[21]
The term "AEGIS" appeared in a short 2003 paper from the Benner Lab which explicitly mentioned a possible connection with extraterrestrial life and NASA. This paper mainly reports on the modification of the HIV reverse transcriptase to incorporate pyDAD (K)-puADA (X) with higher fidelity than earlier solutions. It also claims that AEGIS has been used in FDA-approved commercial tests for HIV and hepatitis C as well as a detection tool for SARS virus but does not mention them by name.[19] The paper was expanded upon by a full length 2004 article.[37] The 2004 article also cites the 1997 research paper for branched DNA assay supported by the Aaron Diamond AIDS Research Center.[38]
Besides interaction with biological enzymes and other systems, the Benner Lab also works to refine the production route and the structure of the bases themselves. For example, the synthesis of Z and P was only reported in 2006[18] and dS was only discovered in 2009.[39]
Benner Lab's research made the headlines in 2019, when it was published under a catchy name "Hachimoji DNA". The paper did not mention AEGIS. The system used a "FAL" variant of T7 RNA polymerase and produced an RNA with secondary structure. It also showed that AEGIS bases do not distort DNA structure outside natural ranges.[40]
Despite the notoriety, the Benner Lab soon stopped using the Hachimoji name, preferring to instead keep their options open among all bases designed for AEGIS. In a 2023 article, the group reports on a way to synthesize DNA with all 12 bases of AEGIS using commercially available enzymes, which would decrease the cost of producing them compared to the chemical method. It also describes a way to apply nanopore sequencing to 12-base DNA.[41]
In a 2024 article from the same laboratory, the concept has been broadened into "anthropogenic evolvable genetic information systems", still with the same acronym.[2]
Remove ads
See also
References
Further reading
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads