Top Qs
Timeline
Chat
Perspective
Messenger RNA
RNA that is read by the ribosome to produce a protein From Wikipedia, the free encyclopedia
Remove ads
Messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
This article needs additional citations for verification. (May 2025) |

mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and the ribosome creates the protein utilizing amino acids carried by transfer RNA (tRNA). This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system.
As in DNA, genetic information in mRNA is contained in the sequence of nucleotides, which are arranged into codons consisting of three ribonucleotides each. Each codon codes for a specific amino acid, except the stop codons, which terminate protein synthesis. The translation of codons into amino acids requires two other types of RNA: transfer RNA, which recognizes the codon and provides the corresponding amino acid, and ribosomal RNA (rRNA), the central component of the ribosome's protein-manufacturing machinery.
The concept of mRNA was first conceived by Sydney Brenner and Francis Crick in 1960 during a conversation with François Jacob. In May 1961, messenger RNA was experimentally characterized in two back-to-back Nature papers: one by Brenner, Jacob, and Meselson, and one by Gros and colleagues (including Watson).[1][2] While analyzing the data in preparation for publication, Jacob and Jacques Monod coined the term "messenger RNA".
Remove ads
Synthesis
Summarize
Perspective
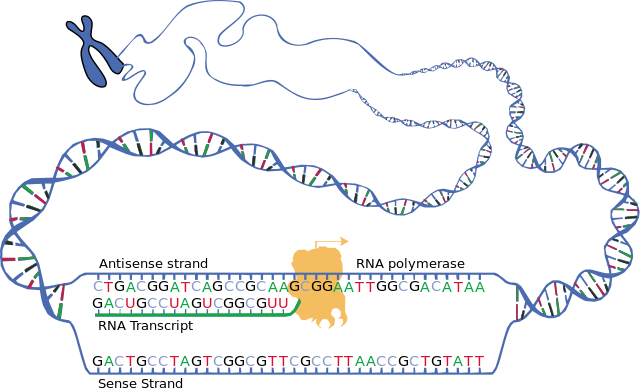
The brief existence of an mRNA molecule begins with transcription, and ultimately ends in degradation. During its life, an mRNA molecule may also be processed, edited, and transported prior to translation. Eukaryotic mRNA molecules often require extensive processing and transport, while prokaryotic mRNA molecules do not. A molecule of eukaryotic mRNA and the proteins surrounding it are together called a messenger RNP[3].
Transcription
Transcription is the process by which genetic information stored in DNA is copied into RNA by the enzyme RNA polymerase.[4] During transcription, RNA polymerase binds to a promoter sequence on the DNA and synthesizes a complementary RNA strand (mRNA) from the DNA template.[5][4]
This process differs between prokaryotes and eukaryotes. In prokaryotes, transcription occurs in the cytoplasm.[5] Because prokaryotes lack a membrane-bound nucleus, ribosomes can attach to the nascent mRNA strand and begin translation while transcription is still in progress.[6]
In eukaryotes, transcription occurs within the cell nucleus.[7] The initial product of transcription is not functional mRNA but is termed precursor mRNA or pre-mRNA.[8] This pre-mRNA must undergo extensive processing (including 5' capping, splicing to remove non-coding introns, and 3' polyadenylation) to become mature mRNA.[8][7] Once processed, the mature mRNA is exported from the nucleus to the cytoplasm for translation.[7]
Uracil substitution for thymine
Whereas DNA contains thymine (T), RNA contains uracil (U). During the process of transcription, the enzyme RNA polymerase incorporates uracil opposite adenine bases located on the DNA template strand. Therefore, the resulting RNA transcript contains uracil in the positions where the coding DNA strand contains thymine.[9][4]
Structurally, uracil–adenine (U–A) base pairs closely resemble thymine–adenine (T–A) base pairs, which ensures that the genetic information carried by the sequence is faithfully preserved.[4]
A frequently cited explanation for the presence of thymine in DNA involves the necessity of genome maintenance. Because cytosine can spontaneously deaminate to form uracil, DNA repair systems recognize uracil as a form of damage. The utilization of thymine as a standard base allows the cell to distinguish legitimate bases from errors, thereby maintaining uracil as a specific signal for repair.[10]
Eukaryotic pre-mRNA processing

Processing of mRNA differs greatly among eukaryotes, bacteria, and archaea. Non-eukaryotic mRNA is, in essence, mature upon transcription and requires no processing, except in rare cases.[11] Eukaryotic pre-mRNA, however, requires several processing steps before its transport to the cytoplasm and its translation by the ribosome.
Splicing
The extensive processing of eukaryotic pre-mRNA that leads to the mature mRNA is the RNA splicing, a mechanism by which introns or outrons (non-coding regions) are removed and exons (coding regions) are joined.[12][13]
5' cap addition

A 5' cap (also termed an RNA cap, an RNA 7-methylguanosine cap, or an RNA m7G cap) is a modified guanine nucleotide that has been added to the "front" or 5' end of a eukaryotic messenger RNA shortly after the start of transcription. The 5' cap consists of a terminal 7-methylguanosine residue that is linked through a 5'-5'-triphosphate bond to the first transcribed nucleotide. Its presence is critical for recognition by the ribosome and protection from RNases.[14]
Cap addition is coupled to transcription, and occurs co-transcriptionally, such that each influences the other. Shortly after the start of transcription, the 5' end of the mRNA being synthesized is bound by a cap-synthesizing complex associated with RNA polymerase. This enzymatic complex catalyzes the chemical reactions that are required for mRNA capping. Synthesis proceeds as a multi-step biochemical reaction.[15]
Editing
In some instances, an mRNA molecule is edited, which changes the nucleotide composition of the transcript. A prominent example in humans involves the apolipoprotein B mRNA. In certain tissues, RNA editing of this transcript creates a premature stop codon, which results in the production of a shorter protein variant. Another well studied mechanism is A-to-I (adenosine-to-inosine) editing. This reaction is catalyzed by ADAR enzymes (adenosine deaminase acting on RNA) and typically occurs within double-stranded RNA regions. A-to-I editing may occur in both coding sequences and untranslated regions. Through these modifications, the process can affect protein recoding, RNA structure, and gene regulation.[16]
Polyadenylation

Polyadenylation is the covalent linkage of a polyadenylyl moiety to a messenger RNA molecule. In eukaryotic organisms most messenger RNA (mRNA) molecules are polyadenylated at the 3' end, but recent studies have shown that short stretches of uridine (oligouridylation) are also common.[17] The poly(A) tail and the protein bound to it aid in protecting mRNA from degradation by exonucleases. Polyadenylation is also important for transcription termination, export of the mRNA from the nucleus, and translation. mRNA can also be polyadenylated in prokaryotic organisms, where poly(A) tails act to facilitate, rather than impede, exonucleolytic degradation.[18] Polyadenylation occurs during and/or immediately after transcription of DNA into RNA. After transcription has been terminated, the mRNA chain is cleaved through the action of an endonuclease complex associated with RNA polymerase. After the mRNA has been cleaved, around 250 adenosine residues are added to the free 3' end at the cleavage site. This reaction is catalyzed by polyadenylate polymerase. Just as in alternative splicing, there can be more than one polyadenylation variant of an mRNA.
Polyadenylation site mutations also occur. The primary RNA transcript of a gene is cleaved at the poly-A addition site, and 100–200 A's are added to the 3' end of the RNA. If this site is altered, an abnormally long and unstable mRNA construct will be formed.
Transport
Another difference between eukaryotes and prokaryotes is mRNA transport. Because eukaryotic transcription and translation is compartmentally separated, eukaryotic mRNAs must be exported from the nucleus to the cytoplasm—a process that may be regulated by different signaling pathways.[19] Mature mRNAs are recognized by their processed modifications and then exported through the nuclear pore by binding to the cap-binding proteins CBP20 and CBP80,[20] as well as the transcription/export complex (TREX).[21][22] Multiple mRNA export pathways have been identified in eukaryotes.[23]
In spatially complex cells, some mRNAs are transported to particular subcellular destinations. In mature neurons, certain mRNA are transported from the soma to dendrites. One site of mRNA translation is at polyribosomes selectively localized beneath synapses.[24] The mRNA for Arc/Arg3.1 is induced by synaptic activity and localizes selectively near active synapses based on signals generated by NMDA receptors.[25] Other mRNAs also move into dendrites in response to external stimuli, such as β-actin mRNA.[26] For export from the nucleus, actin mRNA associates with ZBP1[27] and later with 40S subunit. The complex is bound by a motor protein and is transported to the target location (neurite extension) along the cytoskeleton. Eventually ZBP1 is phosphorylated by Src in order for translation to be initiated.[28] In developing neurons, mRNAs are also transported into growing axons and especially growth cones. Many mRNAs are marked with so-called "zip codes", which target their transport to a specific location.[29][30] mRNAs can also transfer between mammalian cells through structures called tunneling nanotubes.[31][32]
Translation

Because prokaryotic mRNA does not need to be processed or transported, translation by the ribosome can begin immediately after the end of transcription. Therefore, it can be said that prokaryotic translation is coupled to transcription and occurs co-transcriptionally.[33]
In eukaryotic cells the process of translation starts with the information stored in the nucleotide sequence of DNA. This is first transformed into mRNA, then transfer RNA (tRNA) specifies which three-nucleotide codon from the genetic code corresponds to which amino acid.[34]
Eukaryotic mRNA that has been processed and transported to the cytoplasm (i.e., mature mRNA) can then be translated by ribosomes. Translation may occur at ribosomes free in the cytoplasm, or targeted to the endoplasmic reticulum by the signal recognition particle. Therefore, unlike in prokaryotes, eukaryotic translation is not directly coupled to transcription. In some contexts, protein abundance can increase even when mRNA abundance decreases, because translation efficiency and protein turnover are regulated independently of transcript levels; this has been reported for mRNA and protein levels of EEF1A1 in breast cancer.[35][36]
Remove ads
Structure
Summarize
Perspective

Coding regions
Coding regions are composed of codons, which are decoded and translated into proteins by the ribosome; in eukaryotes usually into one and in prokaryotes usually into several. Coding regions begin with the start codon and end with a stop codon. In general, the start codon is an AUG triplet and the stop codon is UAG ("amber"), UAA ("ochre"), or UGA ("opal"). The coding regions tend to be stabilized by internal base pairs; this impedes degradation.[37][38] In addition to being protein-coding, portions of coding regions may serve as regulatory sequences in the pre-mRNA as exonic splicing enhancers or exonic splicing silencers.
Untranslated regions

Untranslated regions (UTRs) are sections of the mRNA before the start codon and after the stop codon that are not translated, termed the five prime untranslated region (5' UTR) and three prime untranslated region (3' UTR), respectively. These regions are transcribed with the coding region and thus are exonic as they are present in the mature mRNA. Several roles in gene expression have been attributed to the untranslated regions, including mRNA stability, mRNA localization, and translational efficiency. The ability of a UTR to perform these functions depends on the sequence of the UTR and can differ between mRNAs. Genetic variants in 3' UTR have also been implicated in disease susceptibility because of the change in RNA structure and protein translation.[39]
The stability of mRNAs may be controlled by the 5' UTR and/or 3' UTR due to varying affinity for RNA degrading enzymes called ribonucleases and for ancillary proteins that can promote or inhibit RNA degradation. (See also, C-rich stability element.)
Translational efficiency, including sometimes the complete inhibition of translation, can be controlled by UTRs. Proteins that bind to either the 3' or 5' UTR may affect translation by influencing the ribosome's ability to bind to the mRNA. MicroRNAs bound to the 3' UTR also may affect translational efficiency or mRNA stability.
Cytoplasmic localization of mRNA is thought to be a function of the 3' UTR. Proteins that are needed in a particular region of the cell can also be translated there; in such a case, the 3' UTR may contain sequences that allow the transcript to be localized to this region for translation.
Some of the elements contained in untranslated regions form a characteristic secondary structure when transcribed into RNA. These structural mRNA elements are involved in regulating the mRNA. Some, such as the SECIS element, are targets for proteins to bind. One class of mRNA element, the riboswitches, directly bind small molecules, changing their fold to modify levels of transcription or translation. In these cases, the mRNA regulates itself.
Poly(A) tail
The 3' poly(A) tail is a long sequence of adenine nucleotides (often several hundred) added to the 3' end of the pre-mRNA. This tail promotes export from the nucleus and translation, and protects the mRNA from degradation.
Monocistronic versus polycistronic mRNA
An mRNA molecule is said to be monocistronic when it contains the genetic information to translate only a single protein chain (polypeptide). This is the case for most of the eukaryotic mRNAs.[40][41] On the other hand, polycistronic mRNA carries several open reading frames (ORFs), each of which is translated into a polypeptide. These polypeptides usually have a related function (they often are the subunits composing a final complex protein) and their coding sequence is grouped and regulated together in a regulatory region, containing a promoter and an operator. Most of the mRNA found in bacteria and archaea is polycistronic,[40] as is the human mitochondrial genome.[42] Dicistronic or bicistronic mRNA encodes only two proteins.
mRNA circularization

In eukaryotes mRNA molecules form circular structures due to an interaction between the eIF4E and poly(A)-binding protein, which both bind to eIF4G, forming an mRNA-protein-mRNA bridge.[43] Circularization is thought to promote cycling of ribosomes on the mRNA leading to time-efficient translation, and may also function to ensure only intact mRNA are translated (partially degraded mRNA characteristically have no m7G cap, or no poly-A tail).[44]
Other mechanisms for circularization exist, particularly in virus mRNA. Poliovirus mRNA uses a cloverleaf section towards its 5' end to bind PCBP2, which binds poly(A)-binding protein, forming the familiar mRNA-protein-mRNA circle. Barley yellow dwarf virus has binding between mRNA segments on its 5' end and 3' end (called kissing stem loops), circularizing the mRNA without any proteins involved.
RNA virus genomes (the + strands of which are translated as mRNA) are also commonly circularized.[45] During genome replication the circularization acts to enhance genome replication speeds, cycling viral RNA-dependent RNA polymerase much the same as the ribosome is hypothesized to cycle.
Remove ads
Degradation
Summarize
Perspective
Different mRNAs within the same cell have distinct lifetimes (stabilities). In bacterial cells, individual mRNAs can survive from seconds to more than an hour. However, the lifetime averages between 1 and 3 minutes, making bacterial mRNA much less stable than eukaryotic mRNA.[46] In mammalian cells, mRNA lifetimes range from several minutes to days.[47] The greater the stability of an mRNA the more protein may be produced from that mRNA. The limited lifetime of mRNA enables a cell to alter protein synthesis rapidly in response to its changing needs. There are many mechanisms that lead to the destruction of an mRNA, some of which are described below.
Prokaryotic mRNA degradation

In general, in prokaryotes the lifetime of mRNA is much shorter than in eukaryotes. Prokaryotes degrade messages by using a combination of ribonucleases, including endonucleases, 3' exonucleases, and 5' exonucleases. In some instances, small RNA molecules (sRNA) tens to hundreds of nucleotides long can stimulate the degradation of specific mRNAs by base-pairing with complementary sequences and facilitating ribonuclease cleavage by RNase III. It was recently shown that bacteria also have a sort of 5' cap consisting of a triphosphate on the 5' end.[48] Removal of two of the phosphates leaves a 5' monophosphate, causing the message to be destroyed by the exonuclease RNase J, which degrades 5' to 3'.
Eukaryotic mRNA turnover
Inside eukaryotic cells, there is a balance between the processes of translation and mRNA decay. Messages that are being actively translated are bound by ribosomes, the eukaryotic initiation factors eIF-4E and eIF-4G, and poly(A)-binding protein. eIF-4E and eIF-4G block the decapping enzyme (DCP2), and poly(A)-binding protein blocks the exosome complex, protecting the ends of the message. The balance between translation and decay is reflected in the size and abundance of cytoplasmic structures known as P-bodies.[49] The poly(A) tail of the mRNA is shortened by specialized exonucleases that are targeted to specific messenger RNAs by a combination of cis-regulatory sequences on the RNA and trans-acting RNA-binding proteins. Poly(A) tail removal is thought to disrupt the circular structure of the message and destabilize the cap binding complex. The message is then subject to degradation by either the exosome complex or the decapping complex. In this way, translationally inactive messages can be destroyed quickly, while active messages remain intact. The mechanism by which translation stops and the message is handed-off to decay complexes is not understood in detail. The majority of mRNA decay was believed to be cytoplasmic; however, recently, a novel mRNA decay pathway was described, which starts in the nucleus.[50]
AU-rich element decay
The presence of AU-rich elements in some mammalian mRNAs tends to destabilize those transcripts through the action of cellular proteins that bind these sequences and stimulate poly(A) tail removal. Loss of the poly(A) tail is thought to promote mRNA degradation by facilitating attack by both the exosome complex[51] and the decapping complex.[52] Rapid mRNA degradation via AU-rich elements is a critical mechanism for preventing the overproduction of potent cytokines such as tumor necrosis factor (TNF) and granulocyte-macrophage colony stimulating factor (GM-CSF).[53] AU-rich elements also regulate the biosynthesis of proto-oncogenic transcription factors like c-Jun and c-Fos.[54]
Nonsense-mediated decay
Eukaryotic messages are subject to surveillance by nonsense-mediated decay (NMD), which checks for the presence of premature stop codons (nonsense codons) in the message. These can arise via incomplete splicing, V(D)J recombination in the adaptive immune system, mutations in DNA, transcription errors, leaky scanning by the ribosome causing a frame shift, and other causes. Detection of a premature stop codon triggers mRNA degradation by 5' decapping, 3' poly(A) tail removal, or endonucleolytic cleavage.[55]
Small interfering RNA (siRNA)
In metazoans, small interfering RNAs (siRNAs) processed by Dicer are incorporated into a complex known as the RNA-induced silencing complex or RISC. This complex contains an endonuclease that cleaves perfectly complementary messages to which the siRNA binds. The resulting mRNA fragments are then destroyed by exonucleases. siRNA is commonly used in laboratories to block the function of genes in cell culture. It is thought to be part of the innate immune system as a defense against double-stranded RNA viruses.[56]
MicroRNA (miRNA)
MicroRNAs (miRNAs) are small RNAs that typically are partially complementary to sequences in metazoan messenger RNAs.[57][58] Binding of a miRNA to a message can repress translation of that message and accelerate poly(A) tail removal, thereby hastening mRNA degradation. The mechanism of action of miRNAs is the subject of active research.[59][60]
Other decay mechanisms
There are other ways by which messages can be degraded, including non-stop decay and silencing by Piwi-interacting RNA (piRNA), among others.
Remove ads
Applications
Summarize
Perspective
The administration of a nucleoside-modified messenger RNA sequence can cause a cell to make a protein, which in turn could directly treat a disease or could function as a vaccine; more indirectly the protein could drive an endogenous stem cell to differentiate in a desired way.[61][62]
The primary challenges of RNA therapy center on delivering the RNA to the appropriate cells.[63] Challenges include the fact that naked RNA sequences naturally degrade after preparation; they may trigger the body's immune system to attack them as an invader; and they are impermeable to the cell membrane.[62] Once within the cell, they must then leave the cell's transport mechanism to take action within the cytoplasm, which houses the necessary ribosomes.[61]
Overcoming these challenges, mRNA as a therapeutic was first put forward in 1989 "after the development of a broadly applicable in vitro transfection technique."[64] In the 1990s, mRNA vaccines for personalized cancer have been developed, relying on non-nucleoside modified mRNA. mRNA based therapies continue to be investigated as a method of treatment or therapy for both cancer as well as auto-immune, metabolic, and respiratory inflammatory diseases. Gene editing therapies such as CRISPR may also benefit from using mRNA to induce cells to make the desired Cas protein.[65]
Since the 2010s, RNA vaccines and other RNA therapeutics have been considered to be "a new class of drugs".[66] The first mRNA-based vaccines received restricted authorization and were rolled out across the world during the COVID-19 pandemic by Pfizer–BioNTech COVID-19 vaccine and Moderna, for example.[67] The 2023 Nobel Prize in Physiology or Medicine was awarded to Katalin Karikó and Drew Weissman for the development of effective mRNA vaccines against COVID-19.[68][69][70] New approaches to modulate RNA levels as a therapeutics include the use of antisense oligonucleotides, including for neurodevelopment diseases associated with high mortality.[71]
Remove ads
History
Summarize
Perspective
Several molecular biology studies during the early 1950s suggested that RNA played a role in protein synthesis, though the specific role remained unclear. For example, in one of the earliest reports, Jacques Monod and his team showed that RNA synthesis was necessary for protein synthesis, specifically during the production of the enzyme β-galactosidase in the bacterium E. coli.[72] Arthur Pardee also found similar RNA accumulation in 1954.[73] In 1953, Alfred Hershey, June Dixon, and Martha Chase studied E. coli infected with bacteriophage T2 and reported that the bacterium's own DNA decreased while the phage's DNA built up inside the infected cells (including DNA containing 5-hydroxymethylcytosine).[74] In hindsight, this has been discussed as part of the chain of observations that led to the concept of mRNA, and it was not recognized at the time as such.[75]
The idea of mRNA was first conceived by Sydney Brenner and Francis Crick on 15 April 1960 at King's College, Cambridge, while François Jacob was telling them about a recent experiment conducted by Arthur Pardee, himself, and Monod (the so-called PaJaMo experiment, which did not prove mRNA existed but suggested the possibility of its existence). With Crick's encouragement, Brenner and Jacob immediately set out to test this new hypothesis, and they contacted Matthew Meselson at the California Institute of Technology for assistance. During the summer of 1960, Brenner, Jacob, and Meselson conducted an experiment in Meselson's laboratory at Caltech which was the first to prove the existence of mRNA. That fall, Jacob and Monod coined the name "messenger RNA" and developed the first theoretical framework to explain its function.[75]
In February 1961, James Watson revealed that his Harvard-based research group had been right behind them with a series of experiments whose results pointed in roughly the same direction. Brenner and the others agreed to Watson's request to delay publication of their research findings. As a result, the Brenner and Watson articles were published simultaneously in the same issue of Nature in May 1961, while that same month, Jacob and Monod published their theoretical framework for mRNA in the Journal of Molecular Biology.[75]
Remove ads
See also
- Extension Poly(A) Test
- GeneCalling, an mRNA profiling technology
- Missense mRNA
- mRNA display
- mRNA surveillance
- Prokaryotic mRNA degradation
- Transcriptome, the sum of all RNA in a cell
- modRNA Nucleoside-modified messenger RNA
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads
