Top Qs
Timeline
Chat
Perspective
PBR322
Artificial plasmid From Wikipedia, the free encyclopedia
Remove ads
pBR322 is a plasmid and was one of the first widely used E. coli cloning vectors. Created in 1977 in the laboratory of Herbert Boyer at the University of California, San Francisco, it was named after Francisco Bolivar Zapata, the postdoctoral researcher and Raymond L. Rodriguez. The p stands for "plasmid," and BR for "Bolivar" and "Rodriguez."

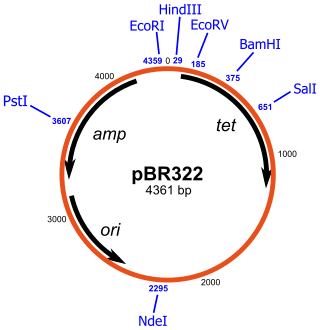
pBR322 is 4361 base pairs in length[1] and has two antibiotic resistance genes – the gene bla encoding the ampicillin resistance (AmpR) protein, and the gene tetA encoding the tetracycline resistance (TetR) protein. It contains the origin of replication of pMB1, and the rop gene, which encodes a restrictor of plasmid copy number. The plasmid has unique restriction sites for more than forty restriction enzymes. Eleven of these forty sites lie within the TetR gene. There are two sites for restriction enzymes HindIII and ClaI within the promoter of the TetR gene. There are six key restriction sites inside the AmpR gene.The source of these antibiotic resistance genes are from pSC101 for Tetracycline and RSF2124 for Ampicillin.[2]
The circular sequence is numbered such that 0 is the middle of the unique EcoRI site and the count increases through the TetR gene. If we have to remove ampicillin for instance, we must use restriction endonuclease or molecular scissors against PstI and then pBR322 will become anti-resistant to ampicillin. The same process of Insertional Inactivation can be applied to Tetracycline. The AmpR gene is penicillin beta-lactamase. Promoters P1 and P3 are for the beta-lactamase gene. P3 is the natural promoter, and P1 is artificially created by the ligation of two different DNA fragments to create pBR322. P2 is in the same region as P1, but it is on the opposite strand and initiates transcription in the direction of the tetracycline resistance gene.[3]
Remove ads
Background
Summarize
Perspective
Early cloning experiments may be conducted using natural plasmids such the ColE1 and pSC101. Each of these plasmids may have its advantages and disadvantages. For example, the ColE1 plasmid and its derivatives have the advantage of higher copy number and allow for chloramphenicol amplification of plasmid to produce a high yield of plasmid, however screening for immunity to colicin E1 is not technically simple.[4] The plasmid pSC101, a natural plasmid from Salmonella panama,[5] confers tetracycline resistance which allows for simpler screening process with antibiotic selection, but it is a low copy number plasmid which does not give a high yield of plasmid. Another plasmid, RSF 2124, which is a derivative of ColE1, confers ampicillin resistance but is larger.
Many other plasmids were artificially constructed to create one that would be ideal for cloning purpose, and pBR322 was found to be most versatile by many and was therefore the one most popularly used.[4] It has two antibiotic resistance genes, as selectable markers, and a number of convenient unique restriction sites that made it suitable as a cloning vector. The plasmid was constructed with genetic material from 3 main sources – the tetracycline resistance gene of pSC101, the ampicillin resistance gene of RSF 2124, and the replication elements of pMB1, a close relative of the ColE1 plasmid.[6][7]
A large number of other plasmids based on pBR322 have since been constructed specifically designed for a wide variety of purposes.[8][9] Examples include the pUC series of plasmids.[10] Most expression vectors for extrachromosomal protein expression and shuttle vectors contain the pBR322 origin of replication, and fragments of pBR322 are very popular in the construction of intraspecies shuttle or binary vectors and vectors for targeted integration and excision of DNA from chromosome.[11]
Remove ads
DNA sequence
Summarize
Perspective
This section is missing information about annotation: what basepairs are what. (March 2019) |
The sequence in pBR322 is[3]
Remove ads
In popular culture
Books
In Michael Crichton's 1990 science fiction novel Jurassic Park “the actual structure of a small fragment of dinosaur DNA“ is shown to visitors to the Jurassic Park Research Institute. In 1992 NCBI Investigator Mark Boguski used BLAST to compare the Jurassic Park sequence with known DNA sequences, and discovered that the book sequence consisted of three sequences from pBR322, one repeated twice, separated by short random sequences. Boguski wrote up his work as a humorous paper, which was published in the journal BioTechniques.[12][13]
See also
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads