Top Qs
Timeline
Chat
Perspective
Slippery sequence
From Wikipedia, the free encyclopedia
Remove ads
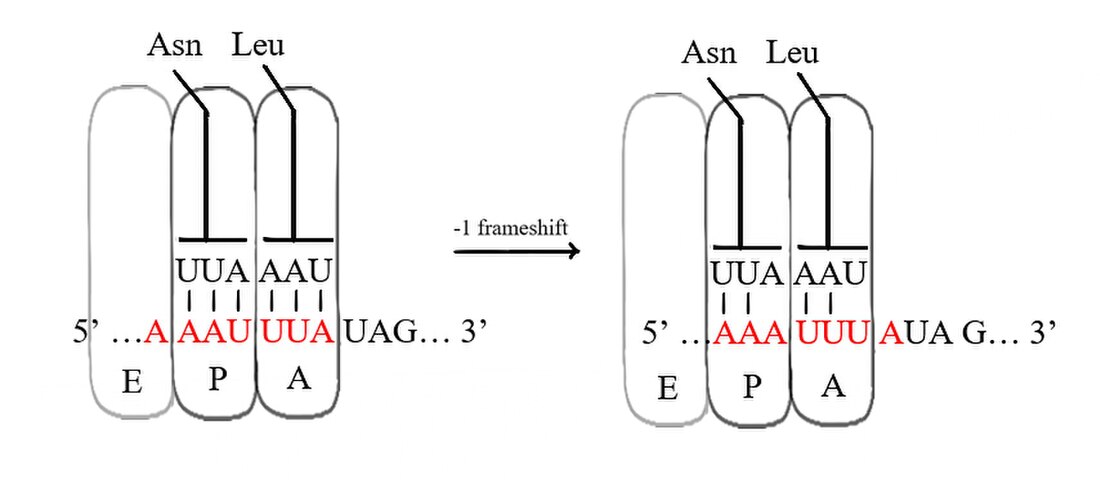
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosome to "slip." This allows a tRNA to shift by 1 base (−1) after it has paired with its anticodon, changing the reading frame.[2][3][4][5][6] A −1 frameshift triggered by such a sequence is a programmed −1 ribosomal frameshift. It is followed by a spacer region, and an RNA secondary structure. Such sequences are common in virus polyproteins.[1]

The frameshift occurs due to wobble pairing. The Gibbs free energy of secondary structures downstream give a hint at how often frameshift happens.[7] Tension on the mRNA molecule also plays a role.[8] A list of slippery sequences found in animal viruses is available from Huang et al.[9]
Slippery sequences that cause a 2-base slip (−2 frameshift) have been constructed out of the HIV UUUUUUA sequence.[8]
Remove ads
See also
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads