Top Qs
Timeline
Chat
Perspective
Spiroplasma
Genus of bacteria From Wikipedia, the free encyclopedia
Remove ads
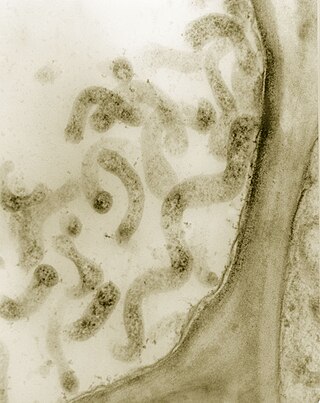
Spiroplasma is a genus of Mollicutes, a group of small bacteria without cell walls. Spiroplasma shares the simple metabolism, parasitic lifestyle, fried-egg colony morphology and small genome of other Mollicutes, but has a distinctive helical morphology, unlike Mycoplasma. It has a spiral shape and moves in a corkscrew motion. Many Spiroplasma are found either in the gut or haemolymph of insects where they can act to manipulate host reproduction, or defend the host as endosymbionts. Spiroplasma are also disease-causing agents in the phloem of plants. Spiroplasmas are fastidious organisms, which require a rich culture medium. Typically they grow well at 30 °C, but not at 37 °C. A few species, notably Spiroplasma mirum, grow well at 37 °C (human body temperature), and cause cataracts and neurological damage in suckling mice.
The best studied species of spiroplasmas are Spiroplasma poulsonii, a reproductive manipulator and defensive insect symbiont, Spiroplasma citri, the causative agent of citrus stubborn disease, and Spiroplasma kunkelii, the causative agent of corn stunt disease.
Remove ads
Genus structure
Summarize
Perspective
Spiroplasma as currently circumscribeed is not monophyletic and consists of four separate clades:
- Spiroplasma sensu stricto consists of the large clade around S. citri. This clade has been subdivided into mirum, chrysopicola, citri, and poulsonii clades, which can be readily distinguished in the phylogenetic trees provided below.
- The ixodetis clade contains two species.
- The apis clade contains 24 species in the broadest view.
- "Candidatus Spiroplasma holothuricola" was named in 2018, creating the fourth clade due to its position on the phylogenetic tree. It was found in an unnamed sea cucumber species close to Zygothuria oxysclera.[2]
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[3] and National Center for Biotechnology Information (NCBI).[4]
| 16S rRNA based LTP_10_2024[5][6][7] | 120 marker proteins based GTDB 09-RS220[8][9][10] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Notes on individual species:
- Spiroplasma mirum has also been called Spiroplasma mira, an attempt at correcting the grammatical gender of the specific epithet to match that of the genus name. However, mirum is of neuter gender and requires no correction. In addition, LPSN (and the LoRN) and GTDB treat S. atrichopogonis as a heterotypic synonym.[11]
- S. insolitum, S. phoeniceum, S. melliferum, and S. diminutum are also grammatically correct and in no need of correction per LPSN.
Remove ads
In arthropods
Summarize
Perspective
Insect symbioses
Many Spiroplasma strains are vertically transmitted endosymbionts of Drosophila species, with a variety of host-altering mechanisms similar to Wolbachia. These strains are from the Spiroplasma poulsonii clade, and can have important effects on host fitness. The S. poulsonii strain of Drosophila neotestacea protects its host against parasitic nematodes. This interaction is an example of defensive symbiosis, where the fitness of the symbiont is intricately tied to the fitness of the host. The D. neotestacea S. poulsonii also defends its fly host from infestation by parasitic wasps.[12][13] The mechanism through which S. poulsonii attacks nematodes and parasitic wasps relies on the presence of toxins called ribosome-inactivating proteins (RIPs), similar to Sarcin or Ricin.[14] These toxins depurinate a conserved adenine site in eukaryotic 28S ribosomal RNA called the Sarcin-Ricin loop by cleaving the N-glycosidic bond between the rRNA backbone and the adenine.[14] Spiroplasma associations highlight a growing movement to consider heritable symbionts as important drivers in patterns of evolution.[15][16] Protection against wasp attack can be thermally sensitive, ablated at lower environmental temperatures.[17][18]
The S. poulsonii strain of Drosophila melanogaster can also attack parasitoid wasps, but is not regarded as a primarily defensive symbiont. This is because this strain called MSRO kills D. melanogaster eggs fertilized by Y-bearing sperm.[19] This mode of reproductive manipulation benefits the symbiont as the female fly has a greater reproductive output than males. Work by Veneti and colleagues demonstrated that male-killing was ablated by loss of function of any gene in the dosage compensation complex (DCC), leading to the hypothesis that the target of male-killing was the single X chromosome of males, and enabled by the DCC binding to this chromosome.[20] Work in D. nebulosa demonstrated male death was associated with widespread apoptosis in male embryos during mid/late embryogenesis.[21] The genetic basis of this male-killing was discovered in 2018, solving a decades-old mystery of how the bacteria targeted male-specific cells.[22] In an interview with the Global Health Institute, Dr. Toshiyuki Harumoto said this discovery is the first example of a bacterial effector protein that affects host cellular machinery in a sex-specific manner, and the first endosymbiont factor identified to explain the cause of male-killing. Thus it should have a big impact on the fields of symbiosis, sex determination, and evolution.[23]
Beyond Drosophila, Spiroplasma sensu stricto and that of the ixodetis clade are also found in many insects and arthropods, including ticks, spiders, bees, ants, beetles, and butterflies:
Crustacean diseases
Crustaceans are an economically important group of arthropods.[39]
- An epidemic of tremor disease in the Chinese mitten crab was traced to a new species which has since been named Spiroplasma eriocheiris.
- S. eriocheiris also causes disease in Procambarus clarkii crayfish.[40]
- S. penaei causes up to 90% mortality in whiteleg shrimps in Colombia.
Remove ads
In plants
Plant diseases
Spiroplasma citri is the causative agent of Citrus stubborn disease, a plant disease affecting species in the genus Citrus.[41] It infects the phloem of the affected plant, causing fruit deformities.
Spiroplasma kunkelii is also referred to as Corn Stunt Spiroplasma as it is the causative agent of Corn stunt disease, a disease of corn and other grasses that stunts plant growth. Spiroplasma kunkelii represents a major economic risk, as corn production in the United States is an industry worth over $50 billion.[42]
Both Spiroplasma citri and Spiroplasma kunkelii are transmitted by leafhoppers.[43][44] Both plant pathogens belong to the citri clade. Another member of the clade that infects plants is S. phoeniceum, which causes periwinkle yellowing disease. The rest of the clade infects arthopods.[36]
Plant symbiosis
Spiroplasma floricola lives on the surface of the flowers of the tulip tree Liriodendron tulipifera.[39]
In vertebrates
Summarize
Perspective
One member of this species, Spiroplasma mirum, readily infects newborn rodents but not adult rodents.[32]
In humans
In 1997, an unnamed species closest to S. taiwanense was found in a newborn with unilateral cataract and anterior uveitis. This is the first known human infection.[45]
In 2014, S. turonicum caused a systemic infection in an immunocompromised individual with hypogammaglobulinemia and rheumatoid arthritis, the latter being treated with biologics. This was the first human systemic infection reported.[46]
In 2022, an unnamed species closest to S. eriocheiris caused a bloodstream and lung infection in a man who underwent surgery for aortic dissection. The genome has been sequenced.[47] GTDB calls this species Spiroplasma sp040940205, a placeholder name based on the GenBank/RefSeq genome assembly identifier.[48]
Transmissible spongiform encephalopathy theory
There is some disputed evidence for the role of spiroplasmas in the etiology of transmissible spongiform encephalopathies (TSEs), due primarily to the work of Frank Bastian, summarized below. Other researchers have failed to replicate this work, while the prion model for TSEs has gained very wide acceptance.[49] A 2006 study appears to refute the role of spiroplasmas in the best small animal scrapie model (hamsters).[50] Bastian et al. (2007) have responded to this challenge with the isolation of a spiroplasma species from scrapie-infected tissue, grown it in cell-free culture, and demonstrated its infectivity in deer. Another experiment in the same study isolates S. mirum from ticks and demonstrates its infectivity in deer. The study also claims S. mirum was previously demonstrated to cause TSE in rodents.[51] A 2011 study fails to cause TSE in raccoons with S. mirum, but succeeded with sick raccoon brain tissue.[52]
In 2014, yet another argument for this theory was put forward by Bastian, this time pointing to the production of alpha-synuclein in mammalian cells cultured with Spiroplasma and biofilm formation. The same article also repeats the previous claims about other supportive evidence.[53] No specific rebuttal has been found among PubMed articles that cite this paper. Only one of the 8 citations dealt with any form of TSE as the main topic.
Remove ads
Genetics and molecular evolution
Spiroplasma, like other mollicutes, have a distinct genetic code, with two rather than three stop codons.[54] Molecular evolution studies, using Spiroplasma passaged vertically in Drosophila, indicate a very fast rate of molecular evolution.[55] Spiroplasma genomes are commonly extremely AT rich, can contain a variety of prophage (viral) elements, and also plasmids.CRISPR defences are found in some members of the genus.[56] Genome sizes are generally between 0.7 and 2.2 Mb.
Remove ads
See also
- Mycoplasma, a similar organism causing disease in animals including humans and linked to autoimmune diseases like rheumatoid arthritis.[57]
- Phytoplasma, another similar organism causing disease in plants.
- Prion
- Virino
- List of bacterial orders
- List of bacteria genera
References
External links
Wikiwand - on
Seamless Wikipedia browsing. On steroids.
Remove ads