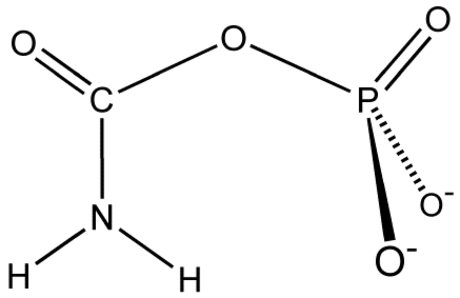
Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine (EC 6.3.5.5) or ammonia (EC 6.3.4.16) and bicarbonate.[1] This ATP-grasp enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
| Carbamoyl phosphate synthetase (ammonia) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 6.3.4.16 | ||||||||
| CAS no. | 37318-69-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||

| CPSase large subunit ATP-binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 the structure of biotin carboxylase, mutant e288k, complexed with atp | |||||||||
| Identifiers | |||||||||
| Symbol | CPSase_L_D2 | ||||||||
| Pfam | PF02786 | ||||||||
| Pfam clan | CL0179 | ||||||||
| InterPro | IPR005479 | ||||||||
| PROSITE | PDOC00676 | ||||||||
| SCOP2 | 1bnc / SCOPe / SUPFAM | ||||||||
| |||||||||
| CPSase large subunit oligomerisation domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of carbamoyl phosphate synthetase complexed with the atp analog amppnp | |||||||||
| Identifiers | |||||||||
| Symbol | CPSase_L_D3 | ||||||||
| Pfam | PF02787 | ||||||||
| InterPro | IPR005480 | ||||||||
| PROSITE | PDOC00676 | ||||||||
| SCOP2 | 1bnc / SCOPe / SUPFAM | ||||||||
| |||||||||
| CPSase large subunit N-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of the biotin carboxylase subunit of pyruvate carboxylase | |||||||||
| Identifiers | |||||||||
| Symbol | CPSase_L_chain | ||||||||
| Pfam | PF00289 | ||||||||
| InterPro | IPR005481 | ||||||||
| PROSITE | PDOC00676 | ||||||||
| SCOP2 | 1bnc / SCOPe / SUPFAM | ||||||||
| |||||||||
| CPSase small subunit N-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 inactivation of the amidotransferase activity of carbamoyl phosphate synthetase by the antibiotic acivicin | |||||||||
| Identifiers | |||||||||
| Symbol | CPSase_sm_chain | ||||||||
| Pfam | PF00988 | ||||||||
| InterPro | IPR002474 | ||||||||
| PROSITE | PDOC00676 | ||||||||
| SCOP2 | 1jdb / SCOPe / SUPFAM | ||||||||
| |||||||||
It represents the first committed step in pyrimidine and arginine biosynthesis in prokaryotes and eukaryotes, and in the urea cycle in most terrestrial vertebrates.[2] Most prokaryotes carry one form of CPSase that participates in both arginine and pyrimidine biosynthesis, however certain bacteria can have separate forms.
There are three different forms that serve very different functions:
- Carbamoyl phosphate synthetase I (mitochondria, urea cycle)
- Carbamoyl phosphate synthetase II (cytosol, pyrimidine metabolism).
- Carbamoyl phosphate synthetase III (found in fish).[3]
Mechanism
Carbamoyl phosphate synthetase has three main steps in its mechanism and is, in essence, irreversible.[4]
- Bicarbonate ion is phosphorylated with ATP to create carboxylphosphate.
- The carboxylphosphate then reacts with ammonia to form carbamic acid, releasing inorganic phosphate.
- A second molecule of ATP then phosphorylates carbamic acid, creating carbamoyl phosphate.
The activity of the enzyme is known to be inhibited by both Tris and HEPES buffers.[5]
Structure
Carbamoyl phosphate synthase (CPSase) is a heterodimeric enzyme composed of a small and a large subunit (with the exception of CPSase III, which is composed of a single polypeptide that may have arisen from gene fusion of the glutaminase and synthetase domains).[2][3][6] CPSase has three active sites, one in the small subunit and two in the large subunit. The small subunit contains the glutamine binding site and catalyses the hydrolysis of glutamine to glutamate and ammonia, which is in turn used by the large chain to synthesize carbamoyl phosphate. The small subunit has a 3-layer beta/beta/alpha structure, and is thought to be mobile in most proteins that carry it. The C-terminal domain of the small subunit of CPSase has glutamine amidotransferase activity. The large subunit has two homologous carboxy phosphate domains, both of which have ATP-binding sites; however, the N-terminal carboxy phosphate domain catalyses the phosphorylation of biocarbonate, while the C-terminal domain catalyses the phosphorylation of the carbamate intermediate.[7] The carboxy phosphate domain found duplicated in the large subunit of CPSase is also present as a single copy in the biotin-dependent enzymes acetyl-CoA carboxylase (ACC), propionyl-CoA carboxylase (PCCase), pyruvate carboxylase (PC) and urea carboxylase.
The large subunit in bacterial CPSase has four structural domains: the carboxy phosphate domain 1, the oligomerisation domain, the carbamoyl phosphate domain 2 and the allosteric domain.[8] CPSase heterodimers from Escherichia coli contain two molecular tunnels: an ammonia tunnel and a carbamate tunnel. These inter-domain tunnels connect the three distinct active sites, and function as conduits for the transport of unstable reaction intermediates (ammonia and carbamate) between successive active sites.[9] The catalytic mechanism of CPSase involves the diffusion of carbamate through the interior of the enzyme from the site of synthesis within the N-terminal domain of the large subunit to the site of phosphorylation within the C-terminal domain.
References
External links
Wikiwand in your browser!
Seamless Wikipedia browsing. On steroids.
Every time you click a link to Wikipedia, Wiktionary or Wikiquote in your browser's search results, it will show the modern Wikiwand interface.
Wikiwand extension is a five stars, simple, with minimum permission required to keep your browsing private, safe and transparent.